 (530) 752-1015
(530) 752-1015
3 Briggs Hall
PFKb
We are studying the function of the family of phosphofructokinase-B (pfkB) type proteins in plants. The Callis lab has begun characterization of two of these members, fructokinase like protein 1 and 2 (FLN1, -2), in Arabidopsis and found both to be important during chloroplast development.
Read MoreRING
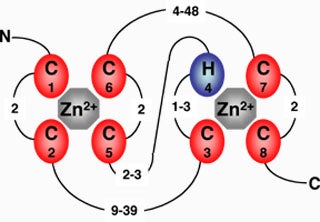
We are examining the activity of another family of proteins containing RING and variant RING domains. These domains have been implicated in the E3 ubiquitin ligase activity of many proteins. We seek to identify putative proteins containing these domains, verify their ligase activity using in vitro ubiquitination assays, identify interacting partners, and characterize their function in vivo by studying the phenotypes of plants with mutations in RING-domain-containing proteins.
Read MoreAuxin Signaling
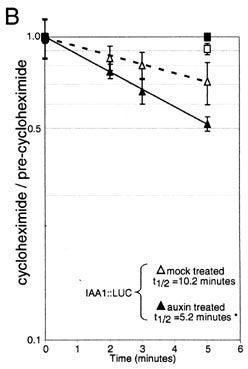
Auxin signal transduction requires the ubiquitin/proteasome pathway. We are interested in studying the degradation of the AUX/IAA family of transcriptional repressors as well as other components of auxin signaling pathways. We hope to characterize the regions required for recognition of the proteolytic targets, and to learn more about the ways in which auxin affects the rate of proteolysis of specific substrates.
Read More

 Join Us On Facebook
Join Us On Facebook Join Us On Twitter
Join Us On Twitter Join Us On In.com
Join Us On In.com Subscribe to RSS
Subscribe to RSS Follow Us On Google+
Follow Us On Google+ Subscribe Us On Youtube
Subscribe Us On Youtube Follow Us On Pinterest
Follow Us On Pinterest Follow Us On Instagram
Follow Us On Instagram Follow Us On Tumblr
Follow Us On Tumblr Subscribe Us On Flickr
Subscribe Us On Flickr